GBIFChecklistHackathon2015Team2Results » History » Revision 7
« Previous |
Revision 7/8
(diff)
| Next »
Andreas Kohlbecker, 06/27/2018 09:13 AM
Web annotation of taxon-level data - Results of Training Hackathon for Checklist Cross-mapping and Precursor National Checklists Generation from GBIF-mediated data¶
Andrea Kohlbecker (leader), Ruud Altenburg, Oskar Kindvall, David Remsen
Sources of biodiversity occurrence data, such as catalogued and indexed by GBIF, may serve as a means to both verify or extend the list of taxa found in national species checklists. They might also serve as the means to start a de-novo national species list. Team 2 focused on a system design that could be used to present assertions of a taxon occurrence within a country - to a presumed expert curator, who might then use their knowledge to assess the assertion and determine whether the taxon should or should not be added to the list. The authoritative Catalogue of Life record - linked through the cross-mapping efforts of Team 1, would then form the record-of-authority for the national list. In addition, negative matches (i.e, species asserted to occur within the country but determined to not belong there - might be linked to a comment or annotation that could serve to inform future users of the GBIF network to the nature of the suspect occurrence. This led to the articulation of the following user story.
User Story 2-1¶
As an owner of a national checklist I want to load my checklist into a system and compare it to the list of taxa assigned to my country within the GBIF index. Matches missing from my national list may 1) represent legitimate missing taxa that should be candidates to add to my list. They may also 2) represent taxa erroneously applied to my country that should be annotated with their suspect status for future users of the record.
Goals¶
- Can the federated GBIF portal be used to support the identification and qualification of novel species occurrence records in the development of national or regional species inventories?
- Can annotation interfaces be used, in combination with authoritative regional or national species lists, to identify and annotate potentially erroneous species occurrences and thus inform future users of GBIF-mobilized data as to this erroneous assessment?
System Design¶
Team 2 came up with the following solutions for each step in the workflow described in Figure 2 below.
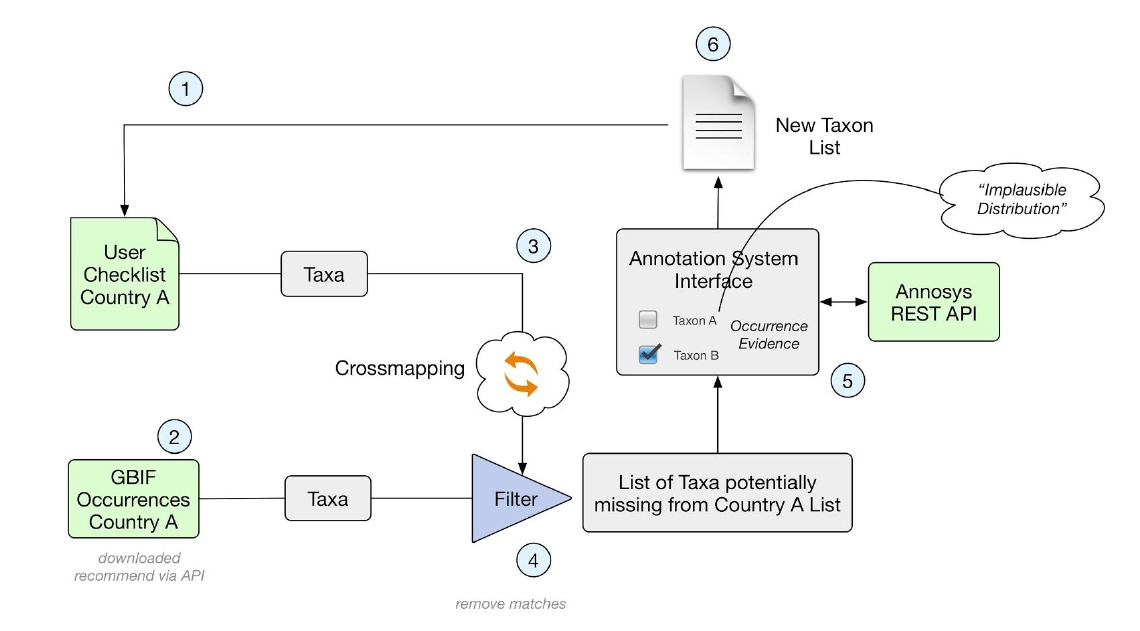
Figure 2. Workflow describing the steps needed to enable comparison between a national checklist and
the taxa represented by GBIF occurrence data.
1) GBIF. The term taxonKey appeared to be a more solid choice.
2) To retrieve the list of taxa represented by occurrence data, the team used SQL distinct selection on speciesKey, scientificName, genus; specificEpithet, infraspecificEpithet. This list was stored in the database (n = 43 387).
3) Cross-matching the checklist with the Catalogue of Life
4) Filtering out the negative matches (the taxa missing from the original national list)
A table was created where all taxa represented by GBIF data was inserted. This table included the following columns: taxonKey, AnnosSysUri, scientificName, blacklisted (bool), taxonStatusGBIF, existsInChecklist, checklistStatus (native, introduced etc), occurrenceRecordCount. The fields existsInChecklist, checklistStatus were updated from the Checklist table. Extraction of the potentially missing taxa was then made by selecting which taxa in the table existsInChecklist is false.
5) Annotate the missing taxa.
a. The team used AnnoSys (https://annosys.bgbm.fu-berlin.de/) to store annotations to the taxon occurrence records. AnnoSys was originally intended to annotate biodiversity occurrence records in ABCD; an XML format. The team extended an Annotation class of AnnoSys so it could handle information about a taxon (as opposed to a taxon occurrence). The team further developed a new annotation model, which is also based on the W3C Open Annotation Data Model (http://www.openannotation.org/spec/core/). General techical documentation and documentation of the open annotation model as used by AnnoSys can be found at http://wiki.bgbm.org/annosys/index.php?title=TechnicalDocumentation
The purpose of the annotation in this case was to express that the distribution for the taxon might, or might not, be correct. In order to express the latter, a interim RDF term (http://rs.gbif.org/terms/ao#ImplausibleDistribution) was introduced.
b. The validation information is then supposed to be posted into AnnoSys using its REST API. We suggest that the annotation should be related to the URL representing the taxon page of GBIF i.e. http://www.gbif.org/species/taxonKey.The annotation should be expressed in a way that should be interpreted as: for the taxon with the taxonKey, all occurrences reported for the specified Country where the establishmentMeans do not clearly indicate non natural occurrence, should be considered as being expected errors.
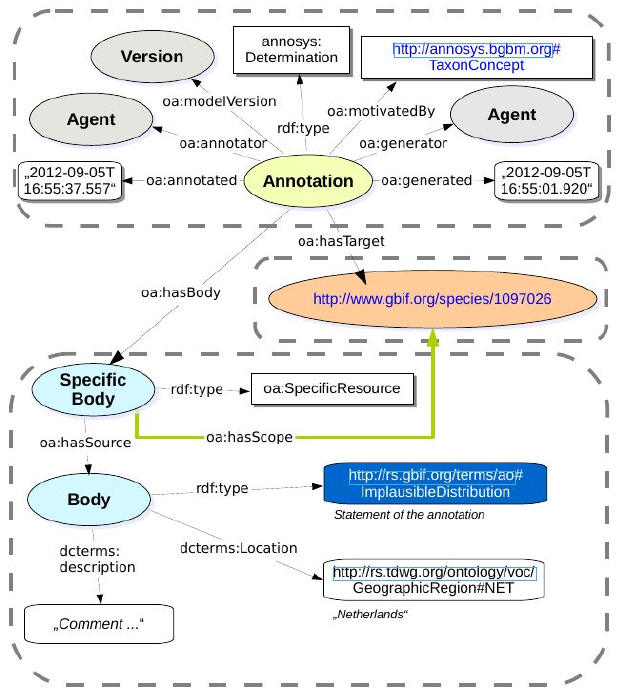
Figure 3 Example PUT request to the AnnoSys to create a new taxon annotation
6) Filter out legitimate missing species candidates for addition to the national list.
The resulting GUI is shown below. It supports annotation of all listed taxa which according to GBIF
occurrence data is likely to candidates to add on to the existing national checklist. For each row a
taxon-specific link to the distribution map generated by GBIF is provided in order to help the user evaluate
the underlying data and judge whether or not the taxon is likely to exist in the country.
To annotate a taxon the user has to use the checkbox. When checked, a taxon-specific annotation is
posted at the AnnoSys repository. When this is done the application posts the response with the URL to
the new annotation in the system database table.
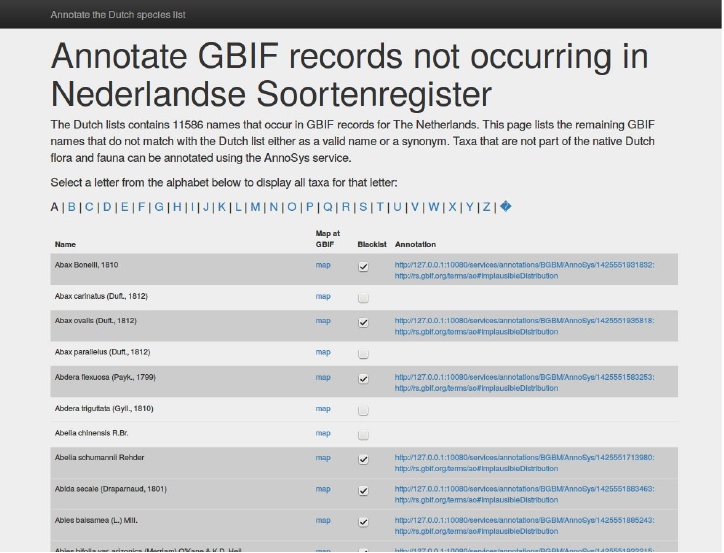
Figure 4. Screen shot of the Annotation GUI with the list taxa potentially missing in the checklist of the Netherlands.
Recommendations¶
As a result of the explorations the team identified an additional user story worthy of further elaboration:
User story 2-2¶
As a curator of GBIF data and harvest processes, I want to be able to look up annotations
indicating that imports of occurrence data with a specified combination Country and Taxon may
be incorrect in order to handle these records in an appropriate way (e.g., records of lions
occurring within the Netherlands and not indicating zoo specimens). They may lead to more
accurate assessment of species composition within countries.
Suggested improvements of existing services¶
In order to make it more feasible to implement a system supporting the User Story investigated here, we
can recognize the need for at least two major improvements of existing web services.
- GBIF occurrence service: A method that can utilize the same set of input parameters for querying occurrence data as the existing that delivers the result as a list of represented (observed) taxa with all relevant DwC terms plus the various GBIF taxonKeys. Relevant could be adding a column for number of occurrence records per taxon listed.
- AnnoSys API: Adding functionality that supports a more generic solution for annotations of information.
Improvements of the preliminary solution¶
The list of potential missing taxa was very long (about 30 000 records). This may partially be explained by
the type of matching: full scientific names including authorship were used. Any spelling variations in the
names of authors would result in non-matching names. Furthermore, the GBIF data was not tested for
semi-invalid data, such as missing lat/lon data (which can be used to verify the location), and an incorrect
country label (data provided in the GBIF DwCA file). After extensive pruning, some values can be used to
score taxa: basisOfRecord, establishmentMeans, type specimen, number of occurrences etc. The
resulting list could be ordered by this score.
A link to the taxon page at GBIF would be useful in the interactive interface in order to support evaluation
of the taxon likelihood of actually being missed in the checklist. That could be if the overall distribution
pattern for that taxon suggest that the natural distribution do not include the target country.
Demo version:http://134.213.149.111/group_2/ (link broken)
Updated by Andreas Kohlbecker almost 6 years ago · 7 revisions