bug #9772
closedImprove DTO usage for character matrix
100%
Description
The character matrix still uses cdm objects for descriptive dataset and descriptions, we need to switch to DTOs as well.
The DTOs should not use cdm objects for creation but pure sql statements.
Discussion on open issues started in #9710. So please see also there for open issues.
Files
Related issues
Updated by Katja Luther over 2 years ago
- Target version changed from Unassigned CDM tickets to Release 5.45
Updated by Katja Luther over 2 years ago
- Status changed from New to In Progress
Now DescriptiveDatasetDto is used and the rowWrapper uses DescriptionDtos as well.
Updated by Andreas Müller over 2 years ago
- Copied from bug #9710: DataIntegrityViolationException after adding a specimen to the matrix added
Updated by Andreas Müller over 2 years ago
Katja Luther wrote:
I also checked the actual nightly build and your test DB and all values are shown.
Now I could reproduce this behaviour, if a second exact value with min/max is added, the type is set to null...
I checked again and found that in the default perspective (Taxonomic) it shows up correctly while in the "Descriptive Dataset" perspective the problem appears. Can you check this?
Updated by Andreas Müller over 2 years ago
I got when saving
last remote method : http://test.e-taxonomy.eu:80/cdmserver/rem_conf_am/remoting/common.service
last remote request client time : 2021-09-20T14:07:25.08
last remote request response header time : Mon, 20 Sep 2021 12:07:25 GMT
client error time : 2021-09-20T14:07:25.127
login : admin
editor version : 5.27.0.202109200833
server : test.e-taxonomy.eu (edit-test) / rem_conf_am
schema version : 5.27.0.0.20210913
os : Windows Server 2012 R2 6.3 amd64
java : 1.8.0_131
org.eclipse.e4.core.di.InjectionException: java.lang.NullPointerException
at org.eclipse.e4.core.internal.di.MethodRequestor.execute(MethodRequestor.java:65)
at org.eclipse.e4.core.internal.di.InjectorImpl.invokeUsingClass(InjectorImpl.java:282)
at org.eclipse.e4.core.internal.di.InjectorImpl.invoke(InjectorImpl.java:247)
at org.eclipse.e4.core.contexts.ContextInjectionFactory.invoke(ContextInjectionFactory.java:90)
at org.eclipse.e4.ui.internal.workbench.PartServiceSaveHandler.save(PartServiceSaveHandler.java:57)
at org.eclipse.ui.internal.WorkbenchWindow$7.save(WorkbenchWindow.java:594)
at org.eclipse.e4.ui.internal.workbench.PartServiceImpl.savePart(PartServiceImpl.java:1390)
at eu.etaxonomy.taxeditor.workbench.SaveHandler.execute(SaveHandler.java:38)
at sun.reflect.NativeMethodAccessorImpl.invoke0(Native Method)
at sun.reflect.NativeMethodAccessorImpl.invoke(NativeMethodAccessorImpl.java:62)
at sun.reflect.DelegatingMethodAccessorImpl.invoke(DelegatingMethodAccessorImpl.java:43)
at java.lang.reflect.Method.invoke(Method.java:498)
at org.eclipse.e4.core.internal.di.MethodRequestor.execute(MethodRequestor.java:55)
at org.eclipse.e4.core.internal.di.InjectorImpl.invokeUsingClass(InjectorImpl.java:282)
at org.eclipse.e4.core.internal.di.InjectorImpl.invoke(InjectorImpl.java:264)
at org.eclipse.e4.core.contexts.ContextInjectionFactory.invoke(ContextInjectionFactory.java:132)
at org.eclipse.e4.core.commands.internal.HandlerServiceHandler.execute(HandlerServiceHandler.java:152)
at org.eclipse.core.commands.Command.executeWithChecks(Command.java:494)
at org.eclipse.core.commands.ParameterizedCommand.executeWithChecks(ParameterizedCommand.java:488)
at org.eclipse.e4.core.commands.internal.HandlerServiceImpl.executeHandler(HandlerServiceImpl.java:210)
at org.eclipse.e4.ui.workbench.renderers.swt.HandledContributionItem.executeItem(HandledContributionItem.java:433)
at org.eclipse.e4.ui.workbench.renderers.swt.AbstractContributionItem.handleWidgetSelection(AbstractContributionItem.java:454)
at org.eclipse.e4.ui.workbench.renderers.swt.AbstractContributionItem$3.handleEvent(AbstractContributionItem.java:482)
at org.eclipse.swt.widgets.EventTable.sendEvent(EventTable.java:84)
at org.eclipse.swt.widgets.Display.sendEvent(Display.java:4418)
at org.eclipse.swt.widgets.Widget.sendEvent(Widget.java:1079)
at org.eclipse.swt.widgets.Display.runDeferredEvents(Display.java:4236)
at org.eclipse.swt.widgets.Display.readAndDispatch(Display.java:3824)
at org.eclipse.e4.ui.internal.workbench.swt.PartRenderingEngine$4.run(PartRenderingEngine.java:1121)
at org.eclipse.core.databinding.observable.Realm.runWithDefault(Realm.java:336)
at org.eclipse.e4.ui.internal.workbench.swt.PartRenderingEngine.run(PartRenderingEngine.java:1022)
at org.eclipse.e4.ui.internal.workbench.E4Workbench.createAndRunUI(E4Workbench.java:150)
at org.eclipse.ui.internal.Workbench$5.run(Workbench.java:693)
at org.eclipse.core.databinding.observable.Realm.runWithDefault(Realm.java:336)
at org.eclipse.ui.internal.Workbench.createAndRunWorkbench(Workbench.java:610)
at org.eclipse.ui.PlatformUI.createAndRunWorkbench(PlatformUI.java:148)
at eu.etaxonomy.taxeditor.Application.start(Application.java:20)
at org.eclipse.equinox.internal.app.EclipseAppHandle.run(EclipseAppHandle.java:196)
at org.eclipse.core.runtime.internal.adaptor.EclipseAppLauncher.runApplication(EclipseAppLauncher.java:134)
at org.eclipse.core.runtime.internal.adaptor.EclipseAppLauncher.start(EclipseAppLauncher.java:104)
at org.eclipse.core.runtime.adaptor.EclipseStarter.run(EclipseStarter.java:388)
at org.eclipse.core.runtime.adaptor.EclipseStarter.run(EclipseStarter.java:243)
at sun.reflect.NativeMethodAccessorImpl.invoke0(Native Method)
at sun.reflect.NativeMethodAccessorImpl.invoke(NativeMethodAccessorImpl.java:62)
at sun.reflect.DelegatingMethodAccessorImpl.invoke(DelegatingMethodAccessorImpl.java:43)
at java.lang.reflect.Method.invoke(Method.java:498)
at org.eclipse.equinox.launcher.Main.invokeFramework(Main.java:673)
at org.eclipse.equinox.launcher.Main.basicRun(Main.java:610)
at org.eclipse.equinox.launcher.Main.run(Main.java:1519)
Caused by: java.lang.NullPointerException
at eu.etaxonomy.cdm.persistence.dto.TermDto.fromTerm(TermDto.java:85)
at eu.etaxonomy.cdm.persistence.dto.TermDto.fromTerm(TermDto.java:73)
at eu.etaxonomy.cdm.api.service.dto.StatisticalMeasurementValueDto.fromStatisticalMeasurementValue(StatisticalMeasurementValueDto.java:34)
at eu.etaxonomy.cdm.api.service.dto.QuantitativeDataDto.fromQuantitativeData(QuantitativeDataDto.java:46)
at eu.etaxonomy.cdm.api.service.dto.DescriptionBaseDto.fromDescription(DescriptionBaseDto.java:249)
at eu.etaxonomy.taxeditor.editor.descriptiveDataSet.matrix.CharacterMatrixPart.save(CharacterMatrixPart.java:249)
at sun.reflect.NativeMethodAccessorImpl.invoke0(Native Method)
at sun.reflect.NativeMethodAccessorImpl.invoke(NativeMethodAccessorImpl.java:62)
at sun.reflect.DelegatingMethodAccessorImpl.invoke(DelegatingMethodAccessorImpl.java:43)
at java.lang.reflect.Method.invoke(Method.java:498)
at org.eclipse.e4.core.internal.di.MethodRequestor.execute(MethodRequestor.java:55)
... 48 more
Updated by Andreas Müller over 2 years ago
Changes in data do currently not get persisted
Updated by Katja Luther over 2 years ago
Andreas Müller wrote:
Katja Luther wrote:
I also checked the actual nightly build and your test DB and all values are shown.
Now I could reproduce this behaviour, if a second exact value with min/max is added, the type is set to null...
I checked again and found that in the default perspective (Taxonomic) it shows up correctly while in the "Descriptive Dataset" perspective the problem appears. Can you check this?
Also with perspective "Descriptive Dataset" and on pesi-hpc I can not reproduce this behaviour...
Updated by Katja Luther over 2 years ago
Andreas Müller wrote:
Changes in data do currently not get persisted
Did you restart the server?
Updated by Andreas Müller over 2 years ago
- Status changed from In Progress to Resolved
- % Done changed from 0 to 30
I put this to resolved as it is for review.
Updated by Andreas Müller over 2 years ago
now the matrix is visible also in DDS perspective again. For whatever reason the problem seems to be solved.
Updated by Andreas Müller over 2 years ago
Katja Luther wrote:
Andreas Müller wrote:
Changes in data do currently not get persisted
Did you restart the server?
AM:
It was restarted after your last cdmlib commit. Ahh, but first time cdmlib did not build. OK, I try again.
Updated by Andreas Müller over 2 years ago
The error changed a bit but I still get an NPE regarding the StatMeasureValues.type:
last remote method : http://test.e-taxonomy.eu:80/cdmserver/rem_conf_am/remoting/progressmonitor.service
last remote request client time : 2021-09-20T15:55:03.484
last remote request response header time : Mon, 20 Sep 2021 13:55:03 GMT
client error time : 2021-09-20T15:55:03.5
login : admin
editor version : 5.27.0.202109201248
server : test.e-taxonomy.eu (edit-test) / rem_conf_am
schema version : 5.27.0.0.20210913
os : Windows Server 2012 R2 6.3 amd64
java : 1.8.0_131
java.lang.NullPointerException
at eu.etaxonomy.cdm.api.service.dto.QuantitativeDataDto.getSpecificStatisticalValue(QuantitativeDataDto.java:76)
at eu.etaxonomy.cdm.api.service.dto.RowWrapperDTO.generateQuantitativeDataString(RowWrapperDTO.java:141)
at eu.etaxonomy.cdm.api.service.dto.RowWrapperDTO.generateDisplayString(RowWrapperDTO.java:133)
at eu.etaxonomy.cdm.api.service.dto.RowWrapperDTO.<init>(RowWrapperDTO.java:61)
at eu.etaxonomy.cdm.api.service.dto.SpecimenRowWrapperDTO.<init>(SpecimenRowWrapperDTO.java:38)
at eu.etaxonomy.cdm.api.service.DescriptiveDataSetService.createSpecimenRowWrapper(DescriptiveDataSetService.java:425)
at eu.etaxonomy.cdm.api.service.DescriptiveDataSetService.createSpecimenRowWrapper(DescriptiveDataSetService.java:432)
at eu.etaxonomy.cdm.api.service.DescriptiveDataSetService.getRowWrapper(DescriptiveDataSetService.java:157)
at sun.reflect.NativeMethodAccessorImpl.invoke0(Native Method)
at sun.reflect.NativeMethodAccessorImpl.invoke(NativeMethodAccessorImpl.java:62)
at sun.reflect.DelegatingMethodAccessorImpl.invoke(DelegatingMethodAccessorImpl.java:43)
at java.lang.reflect.Method.invoke(Method.java:498)
at org.springframework.aop.support.AopUtils.invokeJoinpointUsingReflection(AopUtils.java:333)
at org.springframework.aop.framework.ReflectiveMethodInvocation.invokeJoinpoint(ReflectiveMethodInvocation.java:190)
at org.springframework.aop.framework.ReflectiveMethodInvocation.proceed(ReflectiveMethodInvocation.java:157)
at org.springframework.transaction.interceptor.TransactionInterceptor$1.proceedWithInvocation(TransactionInterceptor.java:99)
at org.springframework.transaction.interceptor.TransactionAspectSupport.invokeWithinTransaction(TransactionAspectSupport.java:283)
at org.springframework.transaction.interceptor.TransactionInterceptor.invoke(TransactionInterceptor.java:96)
at org.springframework.aop.framework.ReflectiveMethodInvocation.proceed(ReflectiveMethodInvocation.java:179)
at org.springframework.aop.framework.JdkDynamicAopProxy.invoke(JdkDynamicAopProxy.java:213)
at com.sun.proxy.$Proxy737.getRowWrapper(Unknown Source)
at eu.etaxonomy.cdm.api.service.longrunningService.LongRunningTasksService$1.doRun(LongRunningTasksService.java:72)
at eu.etaxonomy.cdm.common.monitor.RemotingProgressMonitorThread.run(RemotingProgressMonitorThread.java:42)
Updated by Andreas Müller over 2 years ago
Andreas Müller wrote:
The error changed a bit but I still get an NPE regarding the StatMeasureValues.type:
I checked the above and it is still that type=null data is created. Please check yourself with nightly and search for null values in the DB by
SELECT * FROM StatisticalMeasurementValue smv WHERE smv.type_id IS NULL
after editing StatMeasureValues.
Updated by Andreas Müller over 2 years ago
- Status changed from Resolved to Feedback
There seems to be another issue: when I change only 1 record 8 new records are added in the DB but only 1 should be added. Very strange is that some values are stored which do not appear in the matrix at all (e.g. id=(390,391) in rem_conf_am.StatisticalMeasurementValues with values 445,000 and 2222,0000 which are not visible in the matrix).
Even after only updating a record 4 new records were created. In this case they were all from the same specimen record (in the same line there were 4 StatMeasureValues) but ofcourse the other record should not be created but left as they are. All 4 of them were missing the type_id as mentioned above.
Updated by Andreas Müller over 2 years ago
- Related to feature request #9084: Make it configurable if a Description is deleted only from the Descriptive Data Set or from it's Specimen/Taxon added
Updated by Andreas Müller over 2 years ago
- Target version changed from Release 5.45 to Release 5.27
Updated by Katja Luther over 2 years ago
- Target version changed from Release 5.27 to Release 5.45
Andreas Müller wrote:
There seems to be another issue: when I change only 1 record 8 new records are added in the DB but only 1 should be added. Very strange is that some values are stored which do not appear in the matrix at all (e.g. id=(390,391) in rem_conf_am.StatisticalMeasurementValues with values 445,000 and 2222,0000 which are not visible in the matrix).
Even after only updating a record 4 new records were created. In this case they were all from the same specimen record (in the same line there were 4 StatMeasureValues) but ofcourse the other record should not be created but left as they are. All 4 of them were missing the type_id as mentioned above.
This is because all statisticalMeasurementValues of the updated description are newly created, this is the same as before.
Updated by Katja Luther over 2 years ago
- Target version changed from Release 5.45 to Release 5.27
Updated by Andreas Müller over 2 years ago
The smv.type_id IS NULL issue seems to be solved now. But there is another critical issue: the unit_id is often null for quantitative data though a unit is shown in the popup dialogue for entering data. In the matrix the unit sometimes appears, sometimes it does not. This needs to be checked.
Updated by Katja Luther over 2 years ago
Andreas Müller wrote:
The smv.type_id IS NULL issue seems to be solved now. But there is another critical issue: the unit_id is often null for quantitative data though a unit is shown in the popup dialogue for entering data. In the matrix the unit sometimes appears, sometimes it does not. This needs to be checked.
this is fixed with the next commit.
Updated by Katja Luther over 2 years ago
- Status changed from Feedback to Resolved
- Assignee changed from Katja Luther to Andreas Müller
Save should not create new states or statistical values, but I think this is a new ticket because it is no regression. Therefore this ticket is resolved (NPE and missing unit is solved)
Updated by Katja Luther over 2 years ago
- Related to bug #9781: Merge of QuantitativeData or CategoricalData from Character Matrix should not create new records added
Updated by Andreas Müller over 2 years ago
We will need an update script to try to update existing data (in greece-bupleurum there are 787 quantitative data without unit_id)
Updated by Andreas Müller over 2 years ago
If the unit is changed via the quantitative data editing dialog and persisted this change does not appear in the database
Updated by Andreas Müller over 2 years ago
- Status changed from Resolved to Feedback
- Assignee changed from Andreas Müller to Katja Luther
Updated by Andreas Müller over 2 years ago
- Tags set to additivity
Values are not persisted when entered into a taxon description (literatur description or default description). Or maybe there is another reason. I tested on "AM test dataset with the topmost 3 literatur description and a newly created default description.
This is critical and needs to be fixed before the release.
Updated by Katja Luther over 2 years ago
- Tags deleted (
additivity)
Andreas Müller wrote:
Values are not persisted when entered into a taxon description (literatur description or default description). Or maybe there is another reason. I tested on "AM test dataset with the topmost 3 literatur description and a newly created default description.
This is critical and needs to be fixed before the release.
this is fixed with the last cdmlib commit.
Updated by Andreas Müller over 2 years ago
Katja Luther wrote:
Andreas Müller wrote:
Values are not persisted when entered into a taxon description (literatur description or default description). Or maybe there is another reason. I tested on "AM test dataset with the topmost 3 literatur description and a newly created default description.
This is critical and needs to be fixed before the release.
this is fixed with the last cdmlib commit.
When testing I got
last remote method : http://test.e-taxonomy.eu:80/cdmserver/rem_conf_am/remoting/description.service
last remote request client time : 2021-09-23T13:41:56.755
last remote request response header time : Thu, 23 Sep 2021 11:41:57 GMT
client error time : 2021-09-23T13:41:56.958
login : admin
editor version : 5.27.0.202109230931
server : test.e-taxonomy.eu (edit-test) / rem_conf_am
schema version : 5.27.1.0.20210922
os : Windows Server 2012 R2 6.3 amd64
java : 1.8.0_131
org.eclipse.e4.core.di.InjectionException: java.lang.NullPointerException
at org.eclipse.e4.core.internal.di.MethodRequestor.execute(MethodRequestor.java:65)
at org.eclipse.e4.core.internal.di.InjectorImpl.invokeUsingClass(InjectorImpl.java:282)
...
Caused by: java.lang.NullPointerException
at eu.etaxonomy.cdm.api.service.DescriptionServiceImpl.mergeDescriptions(DescriptionServiceImpl.java:509)
at sun.reflect.NativeMethodAccessorImpl.invoke0(Native Method)
at sun.reflect.NativeMethodAccessorImpl.invoke(NativeMethodAccessorImpl.java:62)
at sun.reflect.DelegatingMethodAccessorImpl.invoke(DelegatingMethodAccessorImpl.java:43)
at java.lang.reflect.Method.invoke(Method.java:498)
at org.springframework.aop.support.AopUtils.invokeJoinpointUsingReflection(AopUtils.java:333)
at org.springframework.aop.framework.ReflectiveMethodInvocation.invokeJoinpoint(ReflectiveMethodInvocation.java:190)
at org.springframework.aop.framework.ReflectiveMethodInvocation.proceed(ReflectiveMethodInvocation.java:157)
at org.springframework.transaction.interceptor.TransactionInterceptor$1.proceedWithInvocation(TransactionInterceptor.java:99)
at org.springframework.transaction.interceptor.TransactionAspectSupport.invokeWithinTransaction(TransactionAspectSupport.java:283)
at org.springframework.transaction.interceptor.TransactionInterceptor.invoke(TransactionInterceptor.java:96)
at org.springframework.aop.framework.ReflectiveMethodInvocation.proceed(ReflectiveMethodInvocation.java:179)
at org.springframework.aop.framework.JdkDynamicAopProxy.invoke(JdkDynamicAopProxy.java:213)
at com.sun.proxy.$Proxy694.mergeDescriptions(Unknown Source)
at sun.reflect.NativeMethodAccessorImpl.invoke0(Native Method)
... 48 more
Updated by Andreas Müller over 2 years ago
- Copied to bug #9785: Update script for missing measurment unit_ids (matrix) added
Updated by Andreas Müller over 2 years ago
Andreas Müller wrote:
We will need an update script to try to update existing data (in greece-bupleurum there are 787 quantitative data without unit_id)
I created a new ticket for this: #9785
Updated by Andreas Müller over 2 years ago
- Tags changed from additivity to additivity, performance
Updated by Katja Luther over 2 years ago
- Tags deleted (
additivity, performance)
Updated by Andreas Müller over 2 years ago
- Tags set to additivity
Currently it looks like quantitative data characters which support >1 measurment units can not be opened in the popup dialogue from the matrix. I tested with "AM test dataset" first character.
Updated by Andreas Müller over 2 years ago
- Tags changed from additivity to additivity, performance
Updated by Katja Luther over 2 years ago
- Tags deleted (
additivity, performance)
Andreas Müller wrote:
Currently it looks like quantitative data characters which support >1 measurment units can not be opened in the popup dialogue from the matrix. I tested with "AM test dataset" first character.
this is fixed.
Updated by Andreas Müller over 2 years ago
- Tags set to additivity, performance
- Status changed from Feedback to Resolved
- Assignee changed from Katja Luther to Andreas Müller
- % Done changed from 30 to 80
All open issues are fixed. Only new issue is the NPE when opening the aggregation. I will check this once the editor was build.
Updated by Katja Luther over 2 years ago
need to be tested:
- add new specimen, add character data -> save , add more character data -> save -> check for doubled entries -> works
- delete character data -> works
- add more than one statistical value -> works
- add literature description -> save -> add data to literature description -> works but sometimes the literature description is added in a new subtree, closing and reopening solves the problem and this problem does not appear if there exist a taxon description already
- add character data to specimen description with default description -> the color change still appears only when the matrix is reloaded. this needs some more work.
- add default description -> save -> add data to default description -> save -> works
- add literature description when editor is dirty -> save -> works
Updated by Andreas Müller over 2 years ago
Katja Luther wrote:
need to be tested:
what does this mean? Did you test it or is it still to be done?
Updated by Katja Luther over 2 years ago
Andreas Müller wrote:
Katja Luther wrote:
need to be tested:
what does this mean? Did you test it or is it still to be done?
I do it right now. If you have other issues, need to be tested, you can add it.
Updated by Katja Luther over 2 years ago
- Status changed from Resolved to In Progress
- Assignee changed from Andreas Müller to Katja Luther
Updated by Katja Luther over 2 years ago
- Status changed from In Progress to Resolved
- Assignee changed from Katja Luther to Andreas Müller
There are still two UI issues, the color of the field where a default description exists, changes (of the color) are only visible after restart the matrix and newly created literature descriptions sometimes show up in a separate subtree.
For the second I don't know if this is a regression, the color issue is, but maybe we can fix it for the next release? What do you think?
Updated by Andreas Müller over 2 years ago
- File picture910-1.png picture910-1.png added
- Status changed from Resolved to Feedback
- Assignee changed from Andreas Müller to Katja Luther
Adding a new literature to a taxon does show the taxon row 2x if a specimen description for this taxon existed before already:
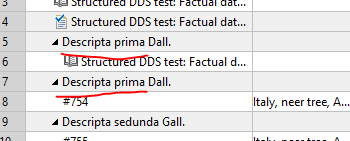
Updated by Andreas Müller over 2 years ago
Andreas Müller wrote:
Adding a new literature to a taxon does show the taxon row 2x if a specimen description for this taxon existed before already:
Upps I just see that this is mentioned in #9772#note-43 already. Sorry.
Is this issue difficult to fix?
Updated by Katja Luther over 2 years ago
Some more tests (with Lactucinae):
found two issues, both are already fixed:
- avoid NPE for quantitative data without measurement unit (for example number of chromosomes)
- before starting aggregation check for changes in matrix.
Updated by Andreas Müller over 2 years ago
This generally seems to work now as expected.
I guess we need to create a couple of follow-up tickets. I could think about the following tickets:
- tests for all functionality related to this ticket and related to the correct handling RowWrapper related issues
- open issues (see commen 43)
- further DTOs to use (create a list of model instances that are still in use for the matrix)
Updated by Andreas Müller over 2 years ago
Can you please also leave a note which model objects have been replaced by this ticket?
And can you check the (new) code, if there is still commented code, unnecessary code and code with unnecessary warnings?
Updated by Andreas Müller over 2 years ago
- Tags changed from additivity, performance to additivity, performance, matrix
- % Done changed from 80 to 90
Generally we can close this ticket.
Updated by Andreas Müller over 2 years ago
Can you also check for other open matrix tickets and add the new tag "matrix" to them so we can check for all open matrix tickets.
Some open issues for the marix might be that there is still the "open in ..." context menu missing which might be very helpful.
Also it would be helpful if the taxon navigator reacts on a selected taxon if the "mit Taxon verknüpfen" button is checked in taxon naviator.
Also it would be great (at least for reviewing) if the records could also show id/uuid of row objects (taxon, specimen). Either in the row at the end (like the bulkeditor) or in the supplementatl data view.
Sooner or later we should also start to implement the details view again to be able to edit some more advanced data like the modifiers. This worked in earlier versions but was removed by PP when implementing a faster version. As we already handle some DTOs in details view I don't think it should be tooo difficult to reenable it. But definetely not a no brainer.
Can you check for existing tickets for all the above issues and create them if not existing?
Updated by Andreas Müller over 2 years ago
Please check also if code is clean. E.g. I found DescriptiveDatasetService.getDescriptiveDataSetDtoByUuid() which includes DAO code like HQL where clauses. This should definetly move to persistence or is there a reason for having in in service layer?
Updated by Andreas Müller over 2 years ago
Same in TaxonNodeServiceImpl.getTaxonNodeDtos().
By the way I wonder if all these HQL statements are necessary. Have you checked if you could use the methods in TaxonNodeFilterDaoHibernateImpl? These methods are well tested and will be maintained in case the TaxonNodeFilter is extended in future. As the TaxonNodeFilter was originally implemented for DDS I guess it could be used at some places instead of using own HQL code. Otherwise we get redundant code for this very complex type of filter which will be difficult to maintain.
But I haven't checked carefully so maybe I am wrong and in the given context the TNfilter can't be used.
Updated by Katja Luther over 2 years ago
Andreas Müller wrote:
Same in TaxonNodeServiceImpl.getTaxonNodeDtos().
By the way I wonder if all these HQL statements are necessary. Have you checked if you could use the methods in TaxonNodeFilterDaoHibernateImpl? These methods are well tested and will be maintained in case the TaxonNodeFilter is extended in future. As the TaxonNodeFilter was originally implemented for DDS I guess it could be used at some places instead of using own HQL code. Otherwise we get redundant code for this very complex type of filter which will be difficult to maintain.
But I haven't checked carefully so maybe I am wrong and in the given context the TNfilter can't be used.
The taxonNodeFilter is used to get the uuids of the filtered taxonnodes, these queries are used to get the dtos for the uuids.
Updated by Katja Luther over 2 years ago
Andreas Müller wrote:
Please check also if code is clean. E.g. I found DescriptiveDatasetService.getDescriptiveDataSetDtoByUuid() which includes DAO code like HQL where clauses. This should definetly move to persistence or is there a reason for having in in service layer?
done
Updated by Katja Luther over 2 years ago
- Status changed from Feedback to Resolved
Code cleaning done for TaxonNodeService/Dao and DescriptiveDatasetService.
Updated by Katja Luther over 2 years ago
Andreas Müller wrote:
Can you please also leave a note which model objects have been replaced by this ticket?
And can you check the (new) code, if there is still commented code, unnecessary code and code with unnecessary warnings?
In the former implementation the DDS was used as cdm object (containing taxonnodes and descriptions) and the descriptions included in rowWrappers where replaced by DTOs
Updated by Katja Luther over 2 years ago
- Related to bug #9800: Open issues for character matrix added
Updated by Andreas Müller over 2 years ago
- Related to task #9842: Improve performance for loading DDS (RowWrapperDTO) added
Updated by Andreas Müller over 2 years ago
- Status changed from Resolved to Closed
There are a couple of open issues (most important probably the loading which uses still model objects and therefore is not performant - #9842 or the creation of new records when records could be updated only).
There are tickets for these open issues so I think we can close this one.
Updated by Katja Luther over 2 years ago
- Related to bug #9885: Write tests for all rowWrapper issues added